tags:
- tuto
- WSL2
- linux
- GPURunning jupyter or IDE on WSL2
First, if you want to install apps through snapd, configure proxy settings in the snap configuration :
(rapids-23.10) brazma@emptre206178:~$ sudo snap set system proxy.http="http://cache.univ-st-etienne.fr:3128"
(rapids-23.10) brazma@emptre206178:~$ sudo snap set system proxy.https="http://cache.univ-st-etienne.fr:3128"
(rapids-23.10) brazma@emptre206178:~$ sudo snap install dataspell --classic
You can then install DataSpell and run a Jupyter server manually following this guide if it doesn't work by default :
Jupyter runs the user's code in a separate process called kernel. The kernel can be a different Python installation (in a different conda environment or virtualenv or Python 2 instead of Python 3) or even an interpreter for a different language (e.g. Julia or R). Kernels are configured by specifying the interpreter and a name and some other parameters (see Jupyter documentation) and configuration can be stored system-wide, for the active environment (or virtualenv) or per user. If nb_conda_kernels is used, additional to statically configured kernels, a separate kernel for each conda environment with ipykernel installed will be available in Jupyter notebooks.
In short, there are three options how to use a conda environment and Jupyter:
Option 1: Run Jupyter server and kernel inside the conda environment
Do something like:
conda create -n my-conda-env # creates new virtual env
conda activate my-conda-env # activate environment in terminal
conda install jupyter # install jupyter + notebook
jupyter notebook # start server + kernel inside my-conda-env
Jupyter will be completely installed in the conda environment. Different versions of Jupyter can be used for different conda environments, but this option might be a bit of overkill. It is enough to include the kernel in the environment, which is the component wrapping Python which runs the code. The rest of Jupyter notebook can be considered as editor or viewer and it is not necessary to install this separately for every environment and include it in every env.yml file. Therefore one of the next two options might be preferable, but this one is the simplest one and definitely fine.
Option 2: Create special kernel for the conda environment
Do something like:
conda create -n my-conda-env # creates new virtual env
conda activate my-conda-env # activate environment in terminal
conda install ipykernel # install Python kernel in new conda env
ipython kernel install --user --name=my-conda-env-kernel # configure Jupyter to use Python kernel
Then run jupyter from the system installation or a different conda environment:
conda deactivate # this step can be omitted by using a different terminal window than before
conda install jupyter # optional, might be installed already in system e.g. by 'apt install jupyter' on debian-based systems
jupyter notebook # run jupyter from system
Name of the kernel and the conda environment are independent from each other, but it might make sense to use a similar name.
Only the Python kernel will be run inside the conda environment, Jupyter from system or a different conda environment will be used - it is not installed in the conda environment. By calling ipython kernel install the jupyter is configured to use the conda environment as kernel, see Jupyter documentation and IPython documentation for more information. In most Linux installations this configuration is a *.json file in ~/.local/share/jupyter/kernels/my-conda-env-kernel/kernel.json:
{
"argv": [
"/opt/miniconda3/envs/my-conda-env/bin/python",
"-m",
"ipykernel_launcher",
"-f",
"{connection_file}"
],
"display_name": "my-conda-env-kernel",
"language": "python"
}
Option 3: Use nb_conda_kernels to use a kernel in the conda environment
When the package nb_conda_kernels is installed, a separate kernel is available automatically for each conda environment containing the conda package ipykernel or a different kernel (R, Julia, ...).
conda activate my-conda-env # this is the environment for your project and code
conda install ipykernel
conda deactivate
conda activate base # could be also some other environment
conda install nb_conda_kernels
jupyter notebook
You should be able to choose the Kernel Python [conda env:my-conda-env]. Note that nb_conda_kernels seems to be available only via conda and not via pip or other package managers like apt.
Running Jupyter in DataSpell
If the auto-starting of a jupyter server on DataSpell doesn't work with an error message along these lines :
[...] Warn(f"{klass} is not importable. Is it installed?", ImportWarning) TypeError: warn() missing 1 required keyword-only argument: 'stacklevel'
Follow these steps :
- Once you managed to run a jupyter server in a terminal following one of the previous options, add a jupyter server in the
Jupyter Serverssettings like so : 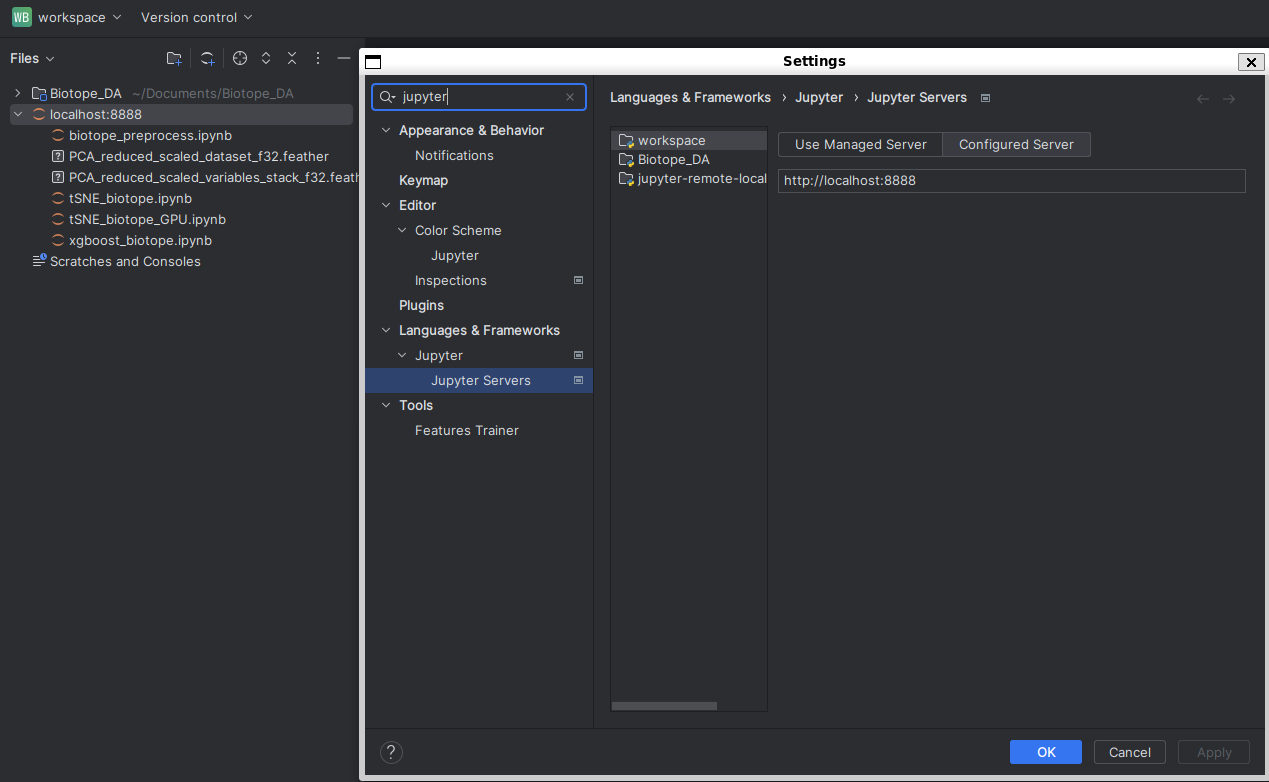
You will see on the left panel the jupyter server being added as it was a project.
Run your files from here instead of the local folder (Biotope_DAhere).
PS : apparently it is possible to run a python environment from the Windows DataSpell using WSL2 interpreter, but i didn't manage to make it work and this is the best working solution so far.
- Once correctly connected, choose the conda environment kernels to run on the rapids-ai conda environment :
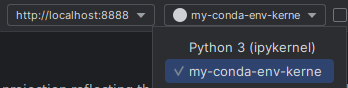
You can check in the bottom right corner which conda environment your code is using, which should be something like rapids-23.10